Grantee Research Project Results
2011 Progress Report: A Multi-scale Dose-response Model of AHR Toxicity Pathway Activation in the Human Liver
EPA Grant Number: R835000Title: A Multi-scale Dose-response Model of AHR Toxicity Pathway Activation in the Human Liver
Investigators: Bhattacharya, Sudin , Andersen, Melvin E. , Clewell, Harvey , McMullen, Patrick
Current Investigators: Bhattacharya, Sudin
Institution: The Hamner Institutes
EPA Project Officer: Chung, Serena
Project Period: June 1, 2011 through May 30, 2015 (Extended to April 30, 2016)
Project Period Covered by this Report: June 1, 2011 through May 31,2012
Project Amount: $750,000
RFA: Computational Toxicology: Biologically-Based Multi-Scale Modeling (2010) RFA Text | Recipients Lists
Research Category: Chemical Safety for Sustainability
Objective:
The overall goal of this project is to develop a multicellular agent-based computational model of the human liver lobule to study the contribution to hepatotoxic injury from multiple levels of biological organization. The activation of the aryl hydrocarbon receptor (AhR) toxicity pathway in hepatocytes exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is used as a case study for data integration and multi-scale model development for mechanistic dose-response prediction.
Progress Summary:
The two main components of this project are: (i) integration of multiple data sources to develop predictive computational models of intracellular toxicity pathways in individual hepatocytes; and (ii) incorporating the single-cell toxicity pathway model into a spatial multicellular “virtual tissue” model of the liver lobule.
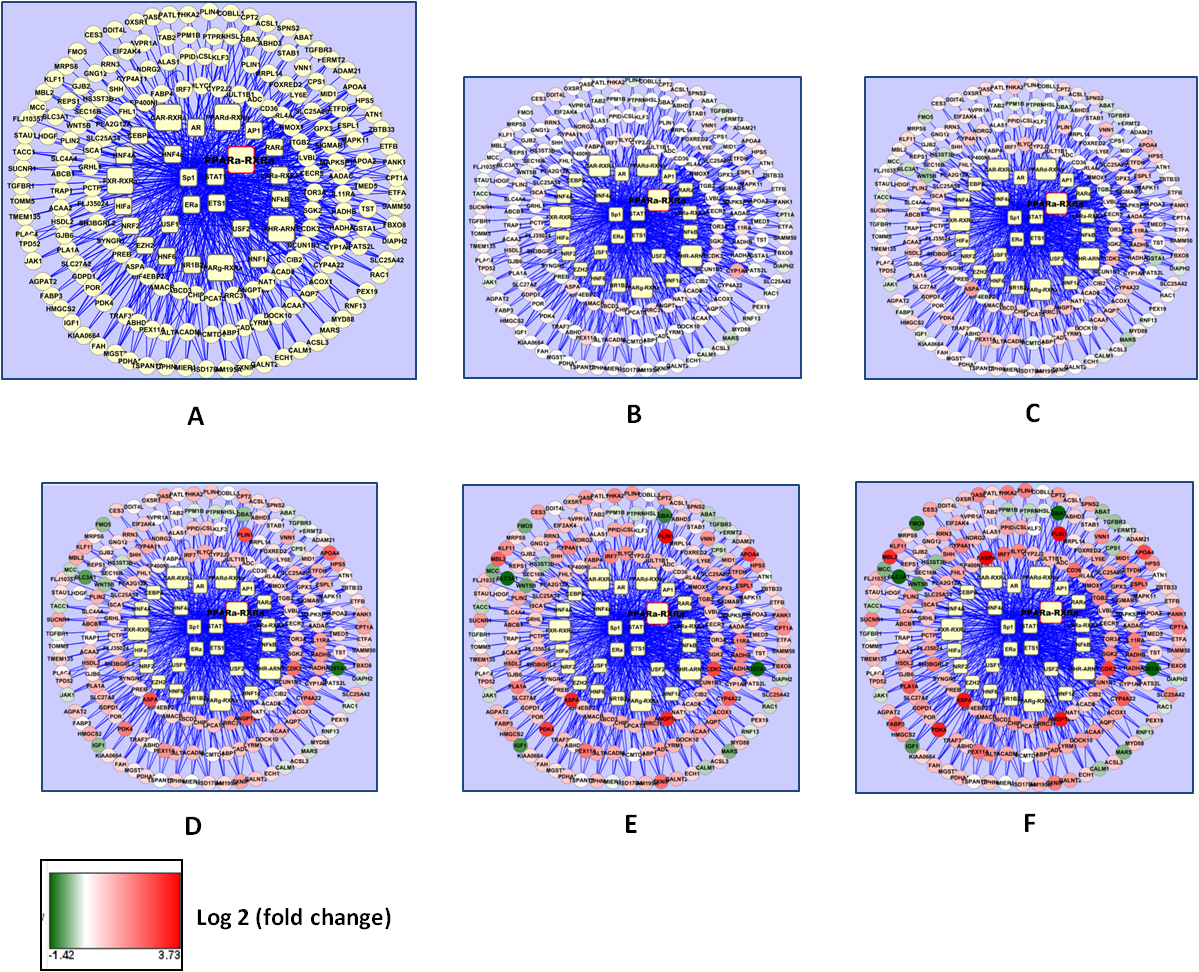
In the first year of the project, we have made significant progress towards the goals related to component (i). We made use of data generated for a different project at The Hamner Institutes on causal network inference for the peroxisome proliferator-activated receptor alpha (PPARα) pathway in primary human hepatocytes. Time- and dose-dependent gene expression data from microarray-based studies was combined with a published ChIP-on-chip analysis of genome-wide PPARα binding and an annotated list of direct PPARα target genes to reconstruct a preliminary transcriptional regulatory network for the PPARα toxicity pathway (Figure 1). This work was presented at the 2011 Wellcome Trust Functional Genomics and Systems Biology meeting. We are currently using the same workflow to reconstruct the transcriptional network for AhR toxicity pathway activation in the liver, the required genome-wide data streams for which are available in the literature.
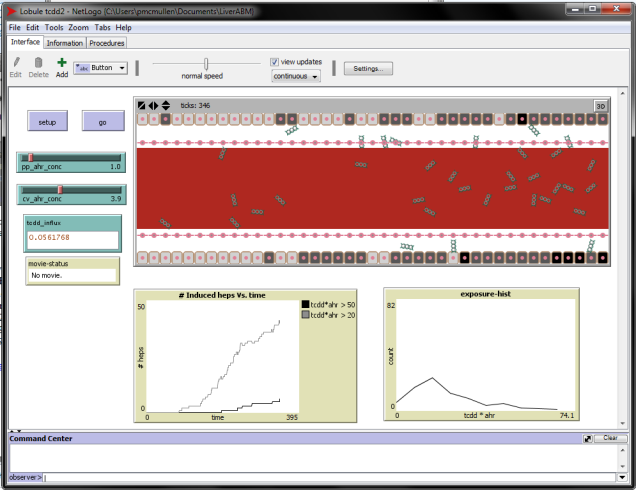
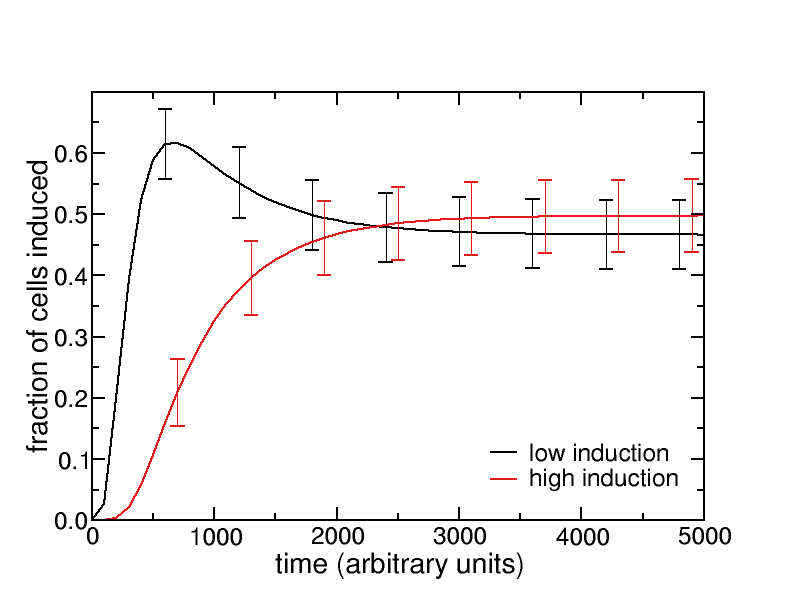
We have taken initial steps towards implementing the multi-scale modeling framework in component (ii) – we now have a working link between Mathematica® (for continuous, equation-based modeling at the organism and intracellular levels) and the agent-based modeling environment NetLogo (for discrete spatial modeling at the tissue level) (Figure 2A). A test-case was simulated for a bolus dose of TCDD administered at the organism-level feeding into the tissue-level agent-based model, with first-order exponential decay in the bulk concentration of TCDD. Enzyme induction in hepatocytes was assumed to be linearly dependent on both the TCDD concentration and intracellular AhR concentration. Simulation of this multi-scale model shows that induction of hepatocytes (qualitatively binned into “low” and “high” induction groups) increases with time before stabilizing (Figure 2B). Because some cells with low induction eventually transition to high induction, the fraction of cells with low induction increases at early times, reaches a maximum, and then decreases.
Figure 1. PPARα transcriptional regulatory network and dose-response. Rectangular nodes indicate regulatory transcription factors (TFs), with the functional PPARα- RXRα heterodimer marked with a bold label. Each edge in the network indicates inferred binding of a TF to the promoter of a target gene (circular nodes). A. The “latent” network. B – F. Network dose-response indicated by expression level of target genes at 72 h for various levels of stimulation with PPARα ligand: B. 0.001 μM; C. 0.01 μM; D. 0.1 μM; E. 1 μM; F. 10 μM.
Figure 2. Multi-scale agent-based model. (A) The NETLOGO agent-based modeling environment. (B) Evolution of hepatocyte induction dynamics with exponential decay of lobular TCDD concentration. Bars indicate ±1 standard deviation over 10,000 simulated replicates.
Future Activities:
The next steps in our work will be to: (i) apply the workflow developed for the PPARα pathway to infer the intra-hepatocyte AhR transcriptional and signaling network; and (ii) incorporate this network into the multicellular agent-based liver lobule model for estimation of tissue-level dose-response.
Journal Articles:
No journal articles submitted with this report: View all 11 publications for this projectSupplemental Keywords:
Computational model, hepatotoxic injury, dose-response, PPARα, 2,3,7,8-tetrachlorodibenzo-p-dioxin, TCDD, aryl hydrocarbon receptor, AhRProgress and Final Reports:
Original AbstractThe perspectives, information and conclusions conveyed in research project abstracts, progress reports, final reports, journal abstracts and journal publications convey the viewpoints of the principal investigator and may not represent the views and policies of ORD and EPA. Conclusions drawn by the principal investigators have not been reviewed by the Agency.